Workflows¶
In Pydra, workflows are DAG of component tasks to be executed on specified inputs. Workflow definitions are dataclasses, which interchangeable with Python and shell tasks definitions and executed in the same way.
Constructor functions¶
Workflows are typically defined using the pydra.compose.workflow.define decorator on a "constructor" function that generates the workflow. For example, given two task definitions, Add and Mul.
[1]:
from pydra.compose import workflow, python
from pydra.utils import show_workflow, print_help
# Example python tasks
@python.define
def Add(a, b):
return a + b
@python.define
def Mul(a, b):
return a * b
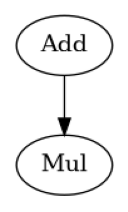
we can create a simple workflow definition using workflow.define to decorate a function that constructs the workflow. Nodes are added to the workflow being constructed by calling workflow.add function.
[2]:
@workflow.define
def BasicWorkflow(a, b):
add = workflow.add(Add(a=a, b=b))
mul = workflow.add(Mul(a=add.out, b=b))
return mul.out
print_help(BasicWorkflow)
show_workflow(BasicWorkflow, figsize=(2, 2.5))
--------------------------------------
Help for Workflow task 'BasicWorkflow'
--------------------------------------
Inputs:
- a: Any
- b: Any
- constructor: Callable[]; default = BasicWorkflow()
Outputs:
- out: Any
workflow.add returns an "outputs" object corresponding to the definition added to the workflow. The fields of the outptus object can be referenced as inputs to downstream workflow nodes. Note that these output fields are just placeholders for the values that will be returned and can’t be used in conditional statements during workflow construction (see Dynamic construction on how to work around this limitation). The fields of the outputs to be
returned by the workflow should be returned in a tuple.
[3]:
from pydra.compose import shell
from fileformats import image, video
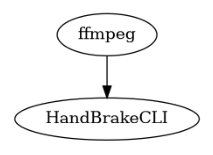
@workflow.define
def ShellWorkflow(
input_video: video.Mp4,
watermark: image.Png,
watermark_dims: tuple[int, int] = (10, 10),
) -> video.Mp4:
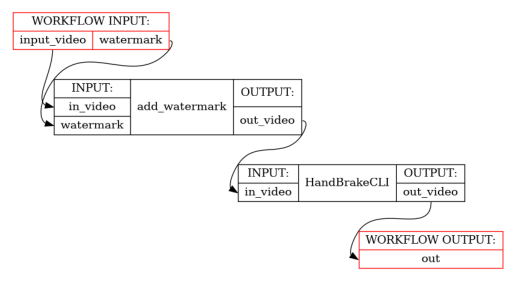
add_watermark = workflow.add(
shell.define(
"ffmpeg -i <in_video> -i <watermark:image/png> "
"-filter_complex <filter> <out|out_video>"
)(
in_video=input_video,
watermark=watermark,
filter="overlay={}:{}".format(*watermark_dims),
)
)
output_video = workflow.add(
shell.define(
"HandBrakeCLI -i <in_video:video/mp4> -o <out|out_video:video/mp4> "
"--width <width:int> --height <height:int>",
)(in_video=add_watermark.out_video, width=1280, height=720)
).out_video
return output_video # test implicit detection of output name
print_help(ShellWorkflow)
show_workflow(ShellWorkflow, figsize=(2.5, 3))
--------------------------------------
Help for Workflow task 'ShellWorkflow'
--------------------------------------
Inputs:
- input_video: video/mp4
- watermark: image/png
- watermark_dims: tuple[int, int]; default = (10, 10)
- constructor: Callable[]; default = ShellWorkflow()
Outputs:
- out: video/mp4
Splitting/combining task inputs¶
Sometimes, you might want to perform the same task over a set of input values/files, and then collect the results into a list to perform further processing. This can be achieved by using the split and combine methods
[4]:
@python.define
def Sum(x: list[float]) -> float:
return sum(x)
@workflow.define
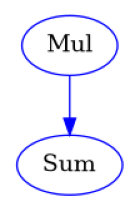
def SplitWorkflow(a: list[int], b: list[float]) -> list[float]:
# Multiply over all combinations of the elements of a and b, then combine the results
# for each a element into a list over each b element
mul = workflow.add(Mul().split(a=a, b=b).combine("a"))
# Sume the multiplications across all all b elements for each a element
sum = workflow.add(Sum(x=mul.out))
return sum.out
print_help(SplitWorkflow)
show_workflow(SplitWorkflow, figsize=(2, 2.5))
--------------------------------------
Help for Workflow task 'SplitWorkflow'
--------------------------------------
Inputs:
- a: list[int]
- b: list[float]
- constructor: Callable[]; default = SplitWorkflow()
Outputs:
- out: list[float]
The combination step doesn’t have to be done on the same step as the split, in which case the splits propagate to downstream nodes
[5]:
@workflow.define
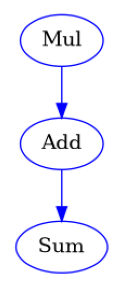
def SplitThenCombineWorkflow(a: list[int], b: list[float], c: float) -> list[float]:
mul = workflow.add(Mul().split(a=a, b=b))
add = workflow.add(Add(a=mul.out, b=c).combine("Mul.a"))
sum = workflow.add(Sum(x=add.out))
return sum.out
print_help(SplitThenCombineWorkflow)
show_workflow(SplitThenCombineWorkflow, figsize=(3, 3.5))
-------------------------------------------------
Help for Workflow task 'SplitThenCombineWorkflow'
-------------------------------------------------
Inputs:
- a: list[int]
- b: list[float]
- c: float
- constructor: Callable[]; default = SplitThenCombineWorkflow()
Outputs:
- out: list[float]
For more advanced discussion on the intricacies of splitting and combining see Splitting and combining
Nested and conditional workflows¶
One of the most powerful features of Pydra is the ability to use inline Python code to conditionally add/omit nodes to workflow, and alter the parameterisation of the nodes, depending on inputs to the workflow
[6]:
@workflow.define
def ConditionalWorkflow(
input_video: video.Mp4,
watermark: image.Png,
watermark_dims: tuple[int, int] | None = None,
) -> video.Mp4:
if watermark_dims is not None:
add_watermark = workflow.add(
shell.define(
"ffmpeg -i <in_video> -i <watermark:image/png> "
"-filter_complex <filter> <out|out_video>"
)(
in_video=input_video,
watermark=watermark,
filter="overlay={}:{}".format(*watermark_dims),
)
)
handbrake_input = add_watermark.out_video
else:
handbrake_input = input_video
output_video = workflow.add(
shell.define(
"HandBrakeCLI -i <in_video:video/mp4> -o <out|out_video:video/mp4> "
"--width <width:int> --height <height:int>",
)(in_video=handbrake_input, width=1280, height=720)
).out_video
return output_video # test implicit detection of output name
print_help(ConditionalWorkflow)
show_workflow(ConditionalWorkflow(watermark_dims=(10, 10)), figsize=(2.5, 3))
--------------------------------------------
Help for Workflow task 'ConditionalWorkflow'
--------------------------------------------
Inputs:
- input_video: video/mp4
- watermark: image/png
- watermark_dims: tuple[int, int] | None; default = None
- constructor: Callable[]; default = ConditionalWorkflow()
Outputs:
- out: video/mp4
Note that outputs of upstream nodes cannot be used in conditional statements, since these are just placeholders at the time the workflow is being constructed. However, you can get around this limitation by placing the conditional logic within a nested workflow
[7]:
@python.define
def Subtract(x: float, y: float) -> float:
return x - y
@workflow.define
def RecursiveNestedWorkflow(a: float, depth: int) -> float:
add = workflow.add(Add(a=a, b=1))
decrement_depth = workflow.add(Subtract(x=depth, y=1))
if depth > 0:
out_node = workflow.add(
RecursiveNestedWorkflow(a=add.out, depth=decrement_depth.out)
)
else:
out_node = add
return out_node.out
print_help(RecursiveNestedWorkflow)
------------------------------------------------
Help for Workflow task 'RecursiveNestedWorkflow'
------------------------------------------------
Inputs:
- a: float
- depth: int
- constructor: Callable[]; default = RecursiveNestedWorkflow()
Outputs:
- out: float
For more detailed discussion of the construction of conditional workflows and "lazy field" placeholders see Conditionals and lazy fields
Type-checking between nodes¶
Pydra utilizes Python type annotations to implement strong type-checking, which is performed when values or upstream outputs are assigned to task inputs.
Job input and output fields do not need to be assigned types, since they will default to typing.Any. However, if they are assigned a type and a value or output from an upstream node conflicts with the type, a TypeError will be raised at construction time.
Note that the type-checking "assumes the best", and will pass if the upstream field is typed by Any or a super-class of the field being assigned to. For example, an input of fileformats.generic.File passed to a field expecting a fileformats.image.Png file type, because Png is a subtype of File, where as fileformats.image.Jpeg input would fail since it is clearly not the intended type.
[8]:
from fileformats import generic
Mp4Handbrake = shell.define(
"HandBrakeCLI -i <in_video:video/mp4> -o <out|out_video:video/mp4> "
"--width <width:int> --height <height:int>",
)
QuicktimeHandbrake = shell.define(
"HandBrakeCLI -i <in_video:video/quicktime> -o <out|out_video:video/quicktime> "
"--width <width:int> --height <height:int>",
)
@workflow.define
def TypeErrorWorkflow(
input_video: video.Mp4,
watermark: generic.File,
watermark_dims: tuple[int, int] = (10, 10),
) -> video.Mp4:
add_watermark = workflow.add(
shell.define(
"ffmpeg -i <in_video> -i <watermark:image/png> "
"-filter_complex <filter> <out|out_video:video/mp4>"
)(
in_video=input_video, # This is OK because in_video is typed Any
watermark=watermark, # Type is OK because generic.File is superclass of image.Png
filter="overlay={}:{}".format(*watermark_dims),
),
name="add_watermark",
)
try:
handbrake = workflow.add(
QuicktimeHandbrake(
in_video=add_watermark.out_video, width=1280, height=720
),
) # This will raise a TypeError because the input video is an Mp4
except TypeError:
handbrake = workflow.add(
Mp4Handbrake(in_video=add_watermark.out_video, width=1280, height=720),
) # The type of the input video is now correct
return handbrake.out_video
print_help(TypeErrorWorkflow)
show_workflow(TypeErrorWorkflow, plot_type="detailed")
------------------------------------------
Help for Workflow task 'TypeErrorWorkflow'
------------------------------------------
Inputs:
- input_video: video/mp4
- watermark: generic/file
- watermark_dims: tuple[int, int]; default = (10, 10)
- constructor: Callable[]; default = TypeErrorWorkflow()
Outputs:
- out: video/mp4
For more detailed discussion on Pydra’s type-checking see Type Checking.
Accessing the workflow object¶
If you need to access the workflow object being constructed from inside the constructor function you can use workflow.this().
[9]:
@python.define(outputs=["divided"])
def Divide(x, y):
return x / y
@workflow.define(outputs=["out1", "out2"])
def DirectAccesWorkflow(a: int, b: float) -> tuple[float, float]:
"""A test workflow demonstration a few alternative ways to set and connect nodes
Args:
a: An integer input
b: A float input
Returns:
out1: The first output
out2: The second output
"""
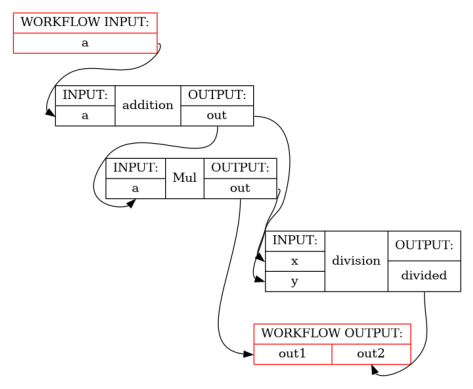
wf = workflow.this()
add = wf.add(Add(a=a, b=b), name="addition")
mul = wf.add(Mul(a=add.out, b=b))
divide = wf.add(Divide(x=wf["addition"].lzout.out, y=mul.out), name="division")
# Alter one of the inputs to a node after it has been initialised
wf["Mul"].inputs.b *= 2
return mul.out, divide.divided
print_help(DirectAccesWorkflow)
show_workflow(DirectAccesWorkflow(b=1), plot_type="detailed")
--------------------------------------------
Help for Workflow task 'DirectAccesWorkflow'
--------------------------------------------
Inputs:
- a: int
An integer input
- b: float
A float input
- constructor: Callable[]; default = DirectAccesWorkflow()
Outputs:
- out1: float
The first output
- out2: float
The second output
Directly access the workflow being constructed also enables you to set the outputs of the workflow directly
[10]:
@workflow.define(outputs={"out1": float, "out2": float})
def SetOutputsOfWorkflow(a: int, b: float):
"""A test workflow demonstration a few alternative ways to set and connect nodes
Args:
a: An integer input
b: A float input
Returns:
out1: The first output
out2: The second output
"""
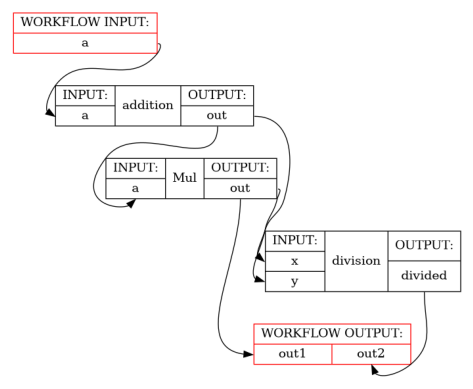
wf = workflow.this()
add = wf.add(Add(a=a, b=b), name="addition")
mul = wf.add(Mul(a=add.out, b=b))
divide = wf.add(Divide(x=wf["addition"].lzout.out, y=mul.out), name="division")
# Alter one of the inputs to a node after it has been initialised
wf["Mul"].inputs.b *= 2
# Set the outputs of the workflow directly
wf.outputs.out1 = mul.out
wf.outputs.out2 = divide.divided
print_help(SetOutputsOfWorkflow)
show_workflow(SetOutputsOfWorkflow(b=3), plot_type="detailed")
---------------------------------------------
Help for Workflow task 'SetOutputsOfWorkflow'
---------------------------------------------
Inputs:
- a: int
An integer input
- b: float
A float input
- constructor: Callable[]; default = SetOutputsOfWorkflow()
Outputs:
- out1: float
The first output
- out2: float
The second output
Setting software environments per node¶
The Advanced execution tutorial showed how the software environment (e.g. Docker container) could be specified for shell tasks by passing the environment variable to the task execution/submission call. For shell tasks within workflows, the software environment used for them is specified when adding a new workflow node, i.e.
[11]:
import tempfile
from pathlib import Path
import numpy as np
from fileformats.medimage import Nifti1
import fileformats.vendor.mrtrix3.medimage as mrtrix3
from pydra.environments import docker
from pydra.compose import workflow, python
from pydra.tasks.mrtrix3.v3_1 import MrConvert, MrThreshold
MRTRIX2NUMPY_DTYPES = {
"Int8": np.dtype("i1"),
"UInt8": np.dtype("u1"),
"Int16LE": np.dtype("<i2"),
"Int16BE": np.dtype(">i2"),
"UInt16LE": np.dtype("<u2"),
"UInt16BE": np.dtype(">u2"),
"Int32LE": np.dtype("<i4"),
"Int32BE": np.dtype(">i4"),
"UInt32LE": np.dtype("<u4"),
"UInt32BE": np.dtype(">u4"),
"Float32LE": np.dtype("<f4"),
"Float32BE": np.dtype(">f4"),
"Float64LE": np.dtype("<f8"),
"Float64BE": np.dtype(">f8"),
"CFloat32LE": np.dtype("<c8"),
"CFloat32BE": np.dtype(">c8"),
"CFloat64LE": np.dtype("<c16"),
"CFloat64BE": np.dtype(">c16"),
}
@workflow.define(outputs=["out_image"])
def ToyMedianThreshold(in_image: Nifti1) -> mrtrix3.ImageFormat:
"""A toy example workflow that
* converts a NIfTI image to MRTrix3 image format with a separate header
* loads the separate data file and selects the median value
"""
input_conversion = workflow.add(
MrConvert(in_file=in_image, out_file="out_file.mih"),
name="input_conversion",
environment=docker.Environment("mrtrix3/mrtrix3", tag="latest"),
)
@python.define
def Median(mih: mrtrix3.ImageHeader) -> float:
"""A bespoke function that reads the separate data file in the MRTrix3 image
header format (i.e. .mih) and calculates the median value.
NB: We could use a MrStats task here, but this is just an example to show how
to use a bespoke function in a workflow.
"""
dtype = MRTRIX2NUMPY_DTYPES[mih.metadata["datatype"].strip()]
data = np.frombuffer(Path.read_bytes(mih.data_file), dtype=dtype)
return np.median(data)
median = workflow.add(Median(mih=input_conversion.out_file))
threshold = workflow.add(
MrThreshold(in_file=in_image, out_file="binary.mif", abs=median.out),
environment=docker.Environment("mrtrix3/mrtrix3", tag="latest"),
)
output_conversion = workflow.add(
MrConvert(in_file=threshold.out_file, out_file="out_image.mif"),
name="output_conversion",
environment=docker.Environment("mrtrix3/mrtrix3", tag="latest"),
)
return output_conversion.out_file
test_dir = Path(tempfile.mkdtemp())
nifti_file = Nifti1.sample(test_dir, seed=0)
wf = ToyMedianThreshold(in_image=nifti_file)
outputs = wf(cache_root=test_dir / "cache")
print(outputs)
ToyMedianThresholdOutputs(out_image=ImageFormat('/tmp/tmp03zx_cp4/cache/workflow-7bc507503c44596cd02a0a6eb59dd71e/out_image.mif'))
See Containers and Environments for more details on how to utilise containers and add support for other software environments.